4. Sites and Trap Types
So far, we have used the same point-type and just a couple of trap-types in our landscape, but MGSurvE can handle complex landscapes with high levels of heterogeneity. We can imagine a landscape with two types of points:
Aquatic Habitats
Blood Haunts
and two types of traps:
Long-range, low-catch (exponential)
Short-range, high-catch (sigmoid)
To setup the landscape, we start by laying down the points with the (x,y) coordinates and now, the last column t as the point-type identifier.
pts = [
[-4.0, 4.00, 0],
[0.25, 8.00, 1],
[5.00, 0.15, 0],
[-1.0, 1.00, 0],
[3.00, 3.00, 1]
]
points = pd.DataFrame(pts, columns=['x', 'y', 't'])
Now, mosquitos don’t pick their destination equally. We can assume for now, that from an aquatic habitat, they preferentially move to a blood haunt, and then back again. We can encode this behavior with the following “mask”:
msk = [
[0.3, 0.7],
[0.7, 0.3]
]
Where the diagonal is the preference towards staying in the same point-type, and the off diagonals encode the probability of moving towards a different point type (in index order of point-types t):
For our traps, we are going to use a similar setup as we did in the previous example:
trp = [
[5.00, 1.00, 0, 0],
[10.0, 0.50, 1, 0],
[10.0, 0.00, 0, 1],
]
traps = pd.DataFrame(trp, columns=['x', 'y', 't', 'f'])
tker = {
0: {'kernel': srv.exponentialDecay, 'params': {'A': 0.4, 'b': 2}},
1: {'kernel': srv.sigmoidDecay, 'params': {'A': .6, 'rate': .5, 'x0': 1}}
}
Where the column t determines the trap kernel to be used by the trap, and the column f if the trap is immovable (1) or movable (0) in the optimization cycle.
Finally, we can setup our landscape as follows:
lnd = srv.Landscape(
points, maskingMatrix=msk, traps=traps, trapsKernels=tker
)
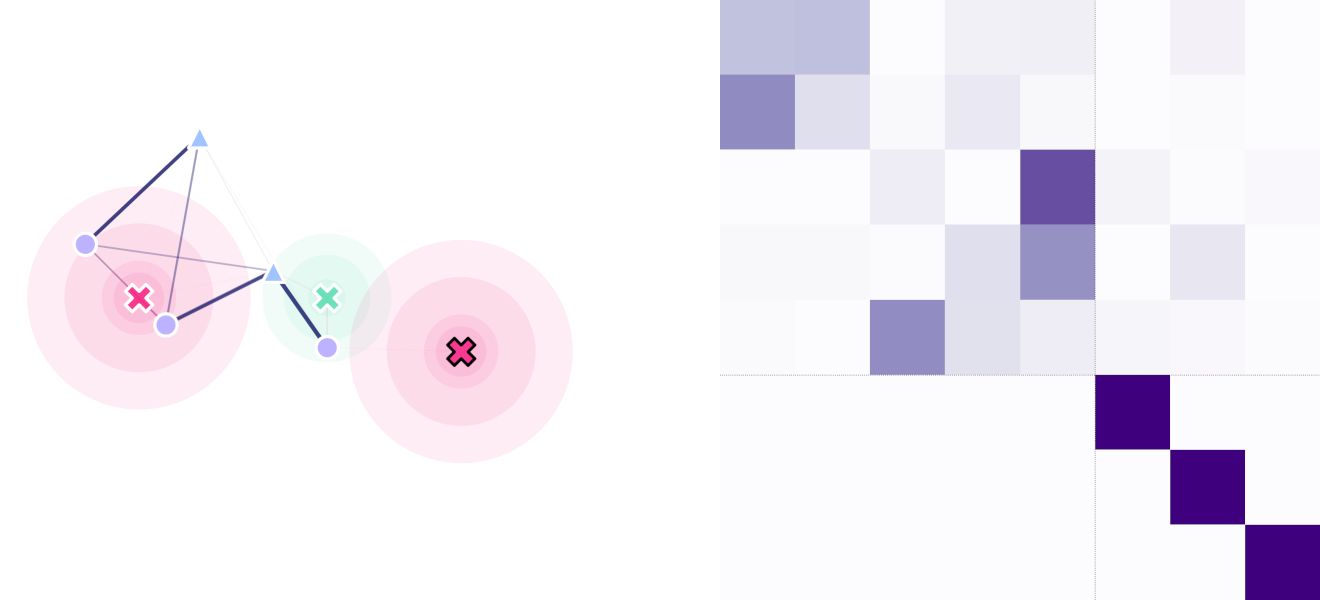
And that’s it! We have our movable sigmoid trap (green), our two exponential-decay traps (magenta), our water sources (circles), and our blood haunts (triangles). We can see that inter-point type transitions are more probable, as defined by our masking matrix.
(fig, ax) = plt.subplots(1, 2, figsize=(15, 15), sharey=False)
lnd.plotSites(fig, ax[0])
lnd.plotMaskedMigrationNetwork(fig, ax[0])
lnd.plotTraps(fig, ax[0])
lnd.plotTrapsNetwork(fig, ax[0])
srv.plotMatrix(fig, ax[1], lnd.trapsMigration, lnd.trapsNumber)
[srv.plotClean(fig, i, frame=False) for i in ax]
The code used for this tutorial can be found in this link.