Pkg Breakdown
MGSurvE is built around a landscape object which stores the information of the sites’ and traps’ positions, along with the mosquito movement information.
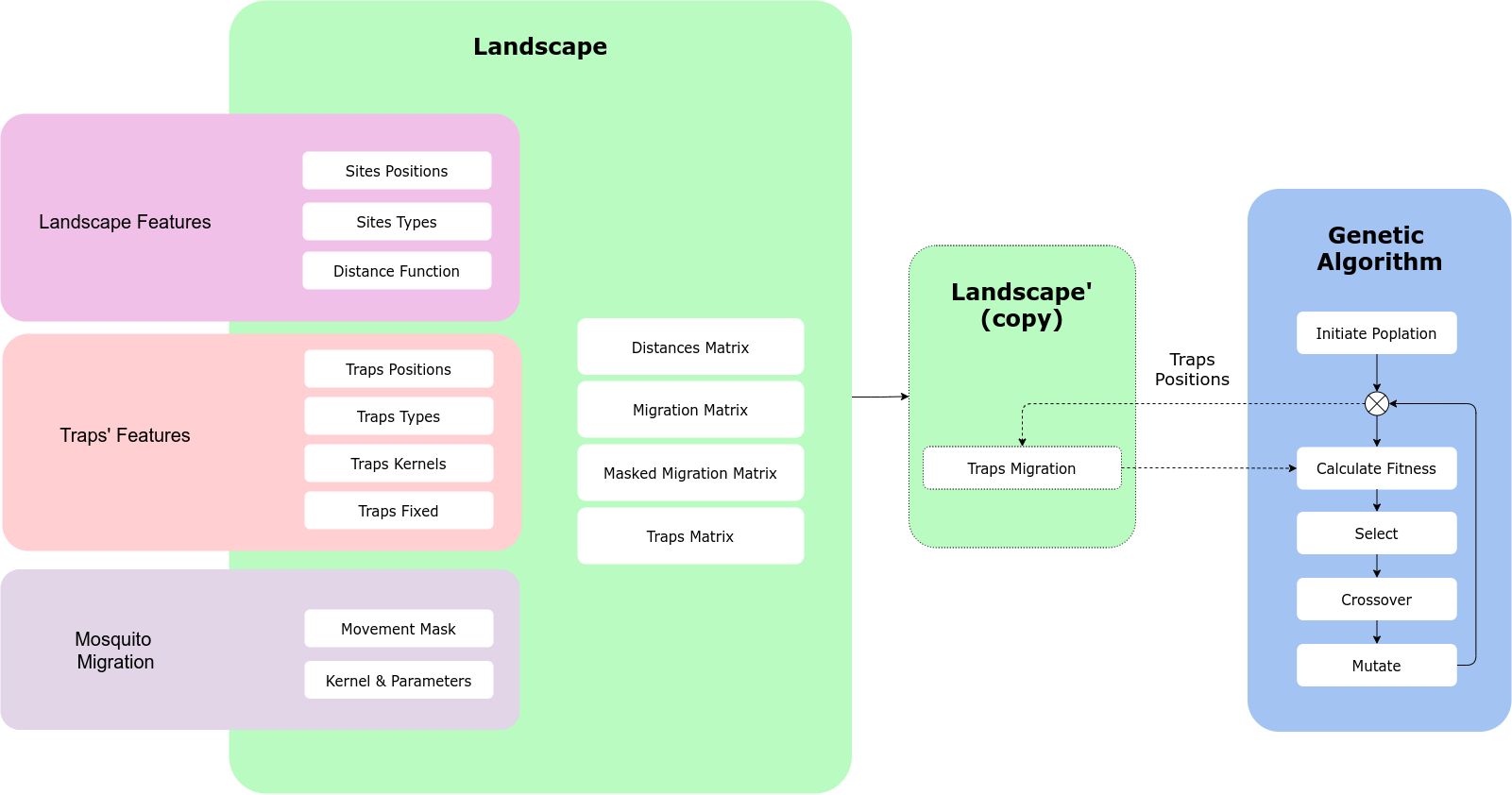
Workflow
A general workflow in MGSurvE looks as follows:
- Define point-set
Positions (pseudo-random or field data)
Types (pseudo-random or field data)
- Define mosquito movement type
Movement kernel (package-provided or custom implementation)
- Define traps’settings
Positions (pseudo-random or field data)
Types (pseudo-random or field data)
Movable/Immovable (for optimization purposes)
Instantiate landscape object with info from previous steps
- Setup DEAP for Genetic-Algorithm (GA) optimization
Register individual’s and population creator
Register mutation operator
Register crossover operator
Register selector operator
Register fitness evaluator
Run the GA optimizer
Update landscape and save results
Components
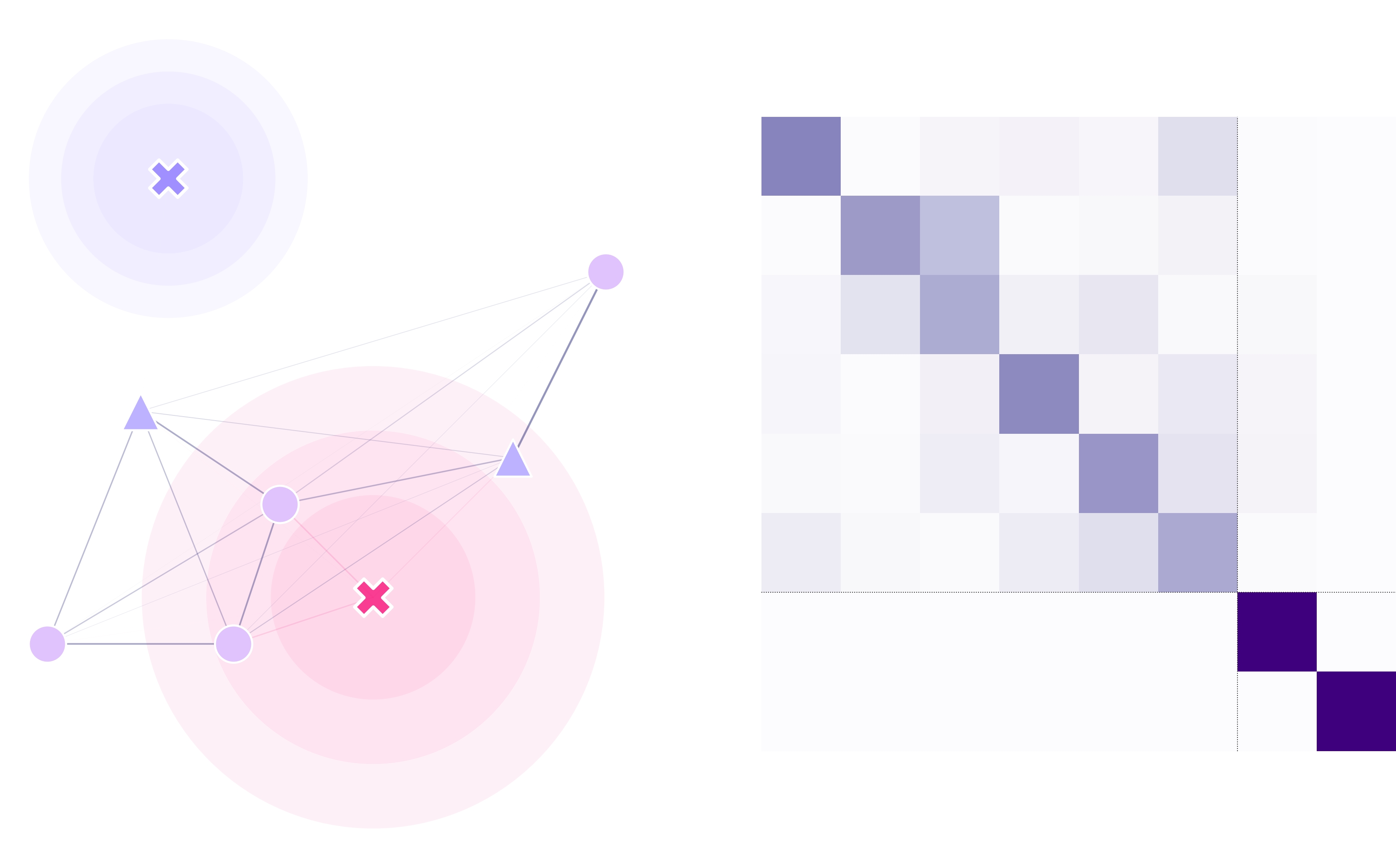
The landscape object contains several elements that make it easy to define, calculate, plot and export our study sites.
Point and Traps Coordinates (lnd.pointCoords, lnd.trapsCoords)
These two numpy arrays contain the information on the locations of points of interest in the landscape. Their ordering matters in terms of how secondary objects are calculated and stored throughout the code (distances and migration matrices preserve the order defined in these structures).
Kernel Function (lnd.kernelFunction)
This function defines the probability for individuals to move from one point to another in a time-step. In their most basic form, they calculate a probability p as a function of the distance d
between points. This kernel function is run to calculate the lnd.migrationMatrix from the lnd.distanceMatrix. It can be defined to take into account other landscape features (such as altitude)
if the parameters are provided (this would not pose a substantial computational burden as it is run only once to setup the landscape).

Traps Kernels (lnd.trapsKernels)
Additional functions that calculate the relation between distance d from the trap to the site from which the individual is moving from, and probability p to fall into a trap (depending on the type of trap to account for attractiveness).
These functions are stored in a dictionary where each entry defines the properties for every trap type.
More complicated functions can be used to account for complex features such as elevation, land-type, and more; but, as these functions need to be run on each iteration of optimization cycles, the more complicated they become, the more processing power that will be required.
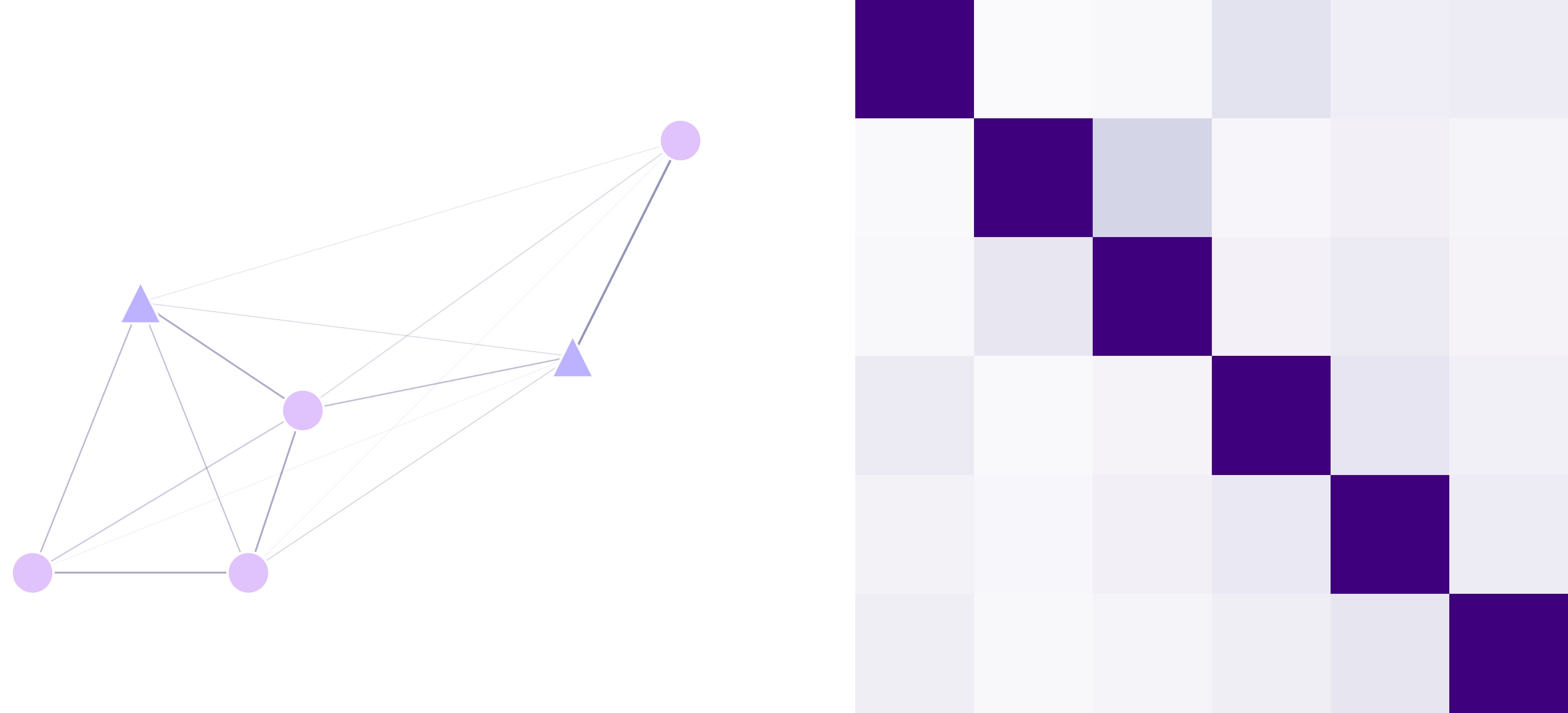
Distances Matrix (lnd.distanceMatrix)
This numpy array contains the distances between all the points in the landscape in the order that they are stored in the
pointCoords. This matrix is calculated point-wise by using the distance function provided to the landscape object.
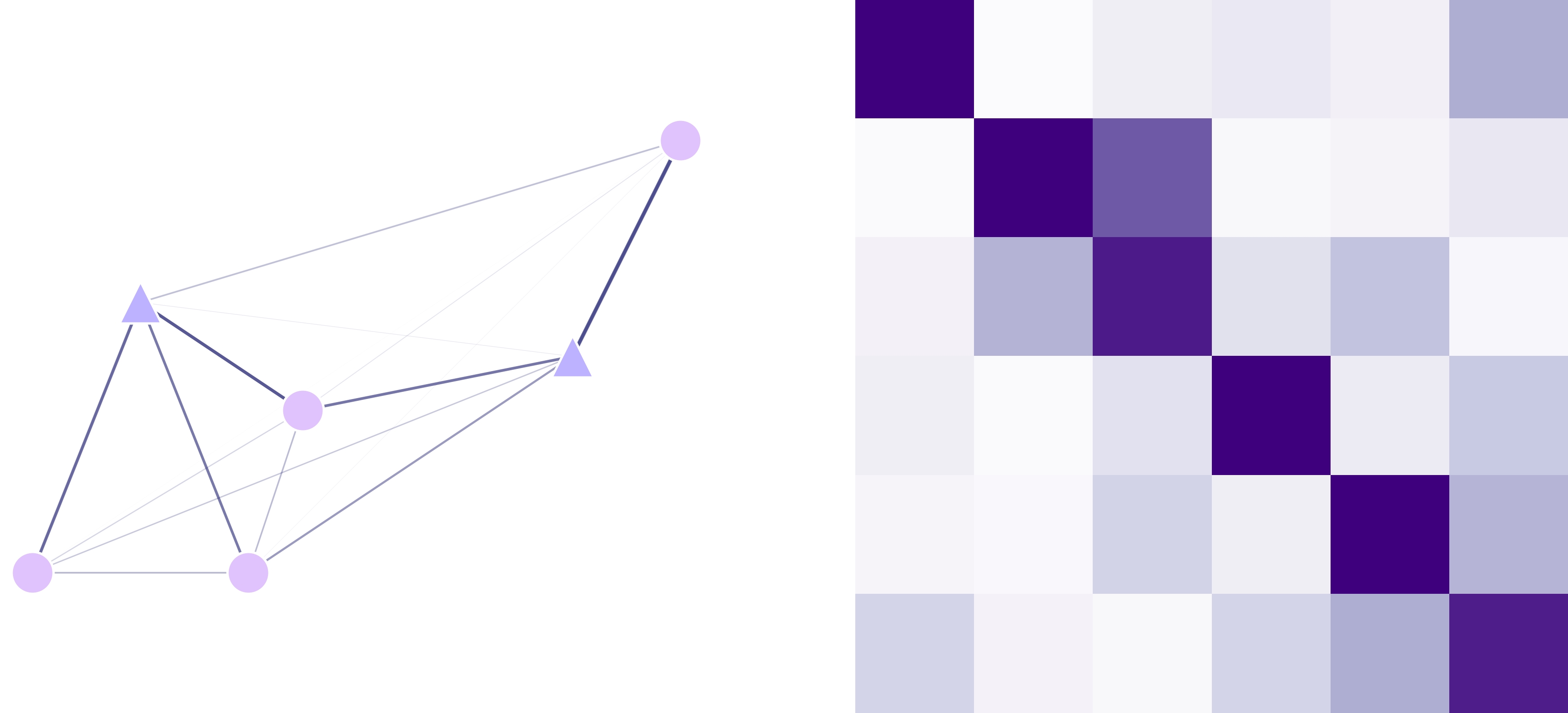
Migration Matrix (lnd.migrationMatrix)
This matrix contains the probabilities of individuals to migrate from point a (row) to point b (column) across
the landscape in a time-step. This matrix is internally calculated using the kernel function and the distance between sites.
Masked Migration Matrix (lnd.maskedMigration)
Similar to the migration matrix but this matrix takes into account the point-types for the probability of movement
(as provided traps mask array). If no traps mask is provided, this matrix is equal to the migration matrix.
Traps Matrix (lnd.trapsMigration)
Finally, the traps matrix contains the probabilities of individuals moving between all the points of the landscape (including
the traps).
Optimization
Some knowledge on basic constrained-optimization operations is needed to make the most of our framework. In particular, on Genetic Algorithms and/or Particle-Swarm Optimization; as these are the two alternatives that are provided out-of-the-box with MGSurvE. Most applications are easy to extend from our tutorials and only require tweaks on the optimization hyper-parameters, but more tailored applications might need some extensions.
Genetic Algorithm (GA)
MGSurvE is designed to integrate into the DEAP Genetic Algorithm framework. We provide some of the basic functions necessary to make the integration as seamless as possible, namely:
initChromosome: to initialize chromosomes for optimization of traps’ positions
x1,y1,x2,y2,...,xn,yn.mutateChromosome: performs a mutation operation but taking into account immovable traps’ flags (an extension of the mutGaussian operator).
mutateChromosomeAsymmetric: an extension of the mutateChromosome that applies two different ranges for mutation’s deviation to account for non-squared landscapes (with significant difference between
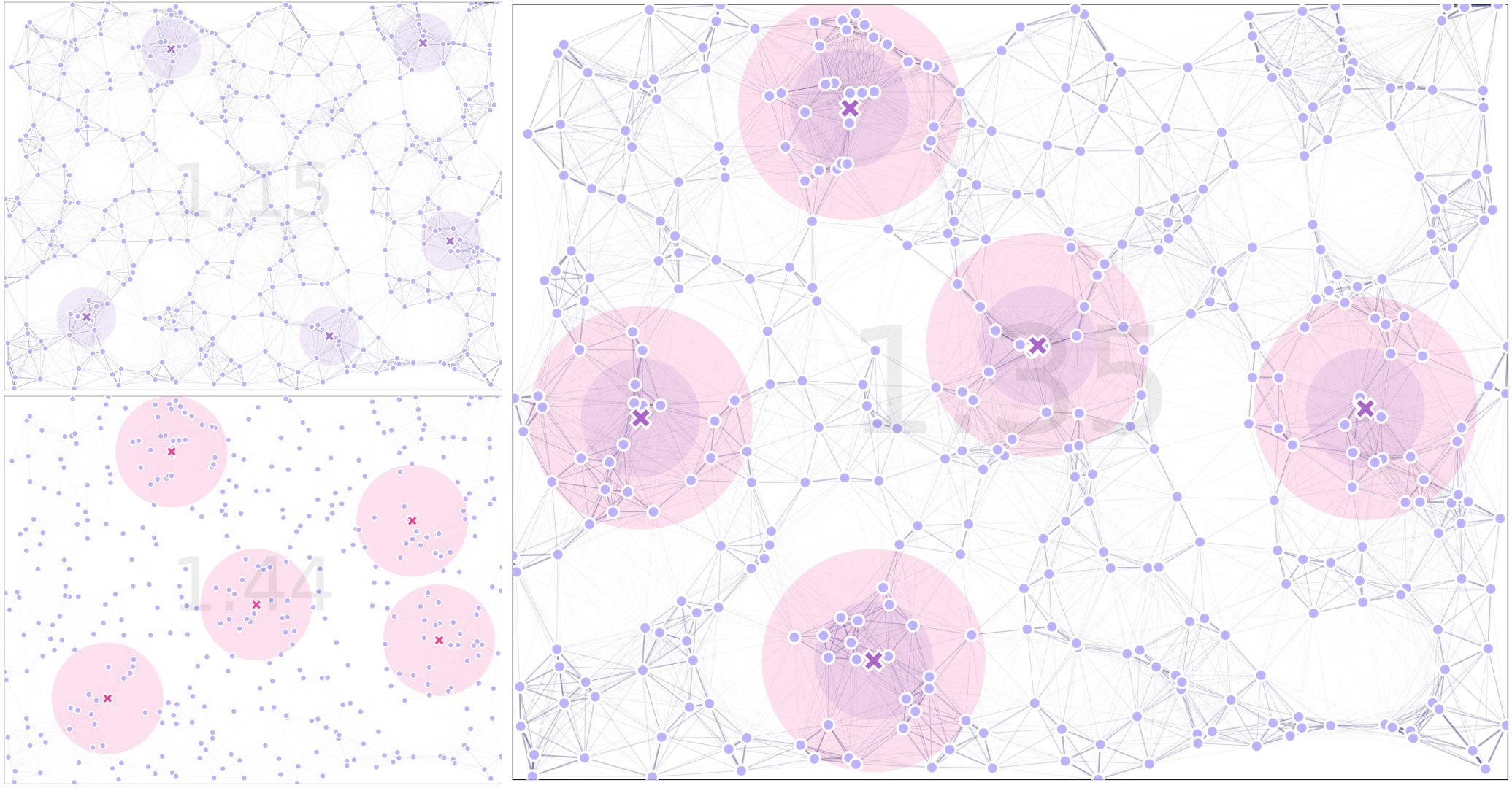
xandyallowed ranges).calcFitness: Calculates the fitness of the traps’ position as defined by Markov’s fundamental matrix to minimize the time it takes for individuals to fall into traps.
calcSexFitness: An extension of calcFitness that allows to give preference to catching one sex over the other if their movement kernels are different.
For a thorough description of the operations, have a look at our examples sections, where we describe how to setup the algorithms for the most common variations of use-cases.
Particle-Swarm Optimization (PSO)
In MGSurvE we also include PSO optimization through the DEAP framework. For PSO, in contrast to GA, we provide a full wrapper object that takes care of all the needed operations within its methods. As such, this alternative is not as flexible as the GA one, but it might prove faster and more stable in certain optimization scenarios.
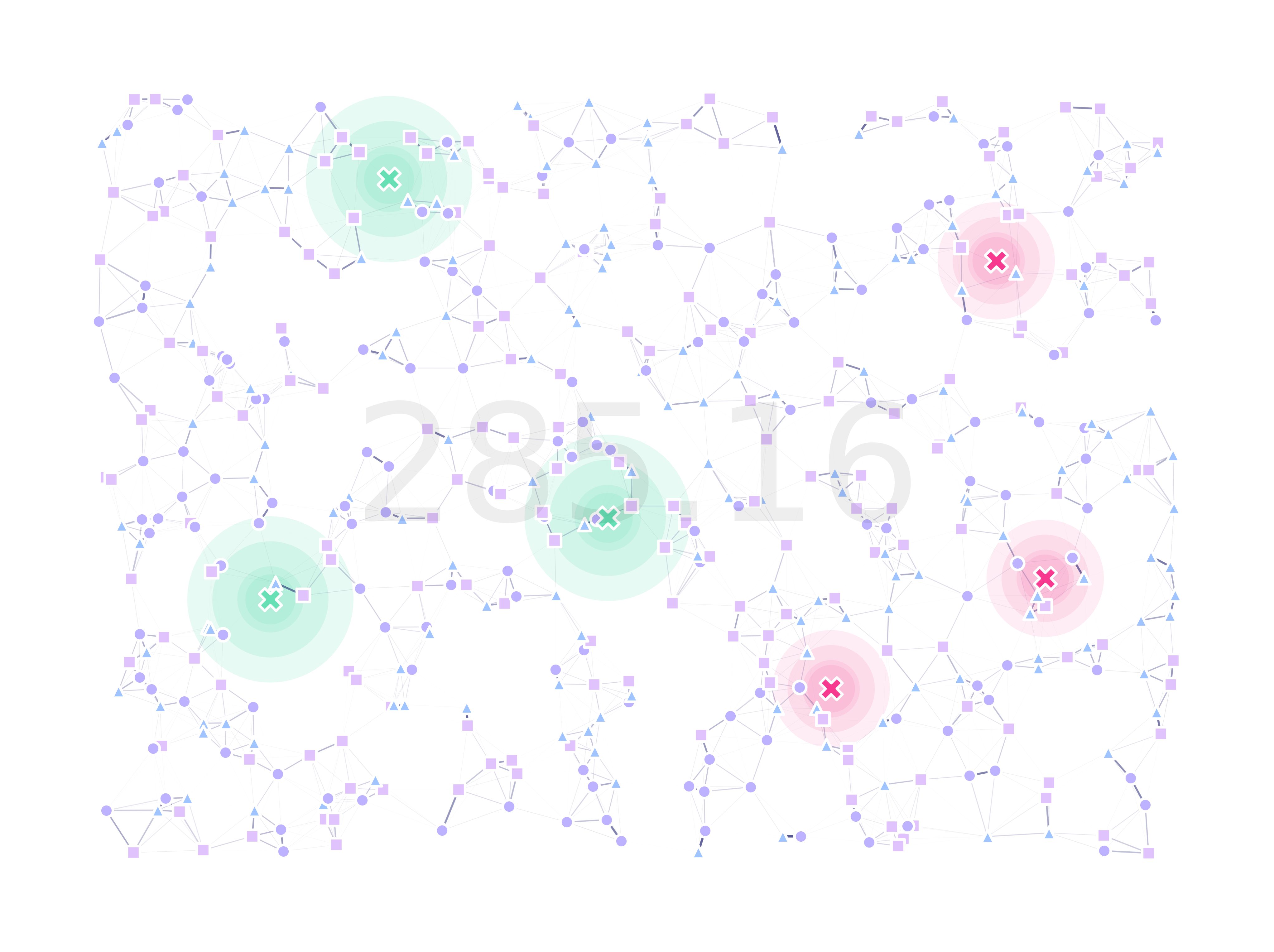
For more information on how to use the particle-swarm alternative to genetic algorithms, have a look at our PSO demo